Chem 112L
Biophysical and Bioanalytical Laboratory
 |
Chem 112L |
 |
Dr. Kalju Kahn
Office: PSB-N 2623
Office hours: Tue 3:30-4:30 PM, "Open Door" policy other times
Phone: 893-6157
E-mail: kalju@chem.ucsb.edu
Website: http://www.chem.ucsb.edu/~kalju
MW2: Adam Pollak (apollak@chem.ucsb.edu) Office hours: Time TBA in Chem 1142 Phone: x8283 TR6: Camille Lawrence (lawrence@lifesci.ucsb.edu) Office hours: Tue 5 PM, Wed 1 PM in Chem 1154 Phone: x5845
Chem 112L is an undergraduate laboratory course where students use advanced instrumental techniques to study the structure and function of biological molecules. Students will work on seven projects and learn to use techniques such as circular dichroism, nuclear magnetic resonance spectroscopy, biological mass spectrometry, differential UV spectrophotometry, protein crystallography, and computational biochemistry.
Lab section 1: Mon 2:00-5:50; Wed 2:00-5:50;
Lab section 2: Tue 6:00-9:50; Thu 6:00-9:50;
Lab sections are in PSB-N 2619 or Chem 1153 (Computer lab)
| Syllabus | General information about the course | |
| Schedule | Course schedule: MW2 (2012) | |
| Schedule | Course schedule: TR6 (2012) | |
| History | Course website for Chem 112L (Spring 2011) | Link |
| Grading | Grading Rubrics | DOC |
| Upload | Submit your assignments as PDF files | Link |
| Exams | How to prepare for the first midterm | |
| Exams | First Midterm, Spring 2011 | |
| Exams | Selected CHEM 112C Lecture Materials | ZIP |
| Exams | Hydrogen Atom: Solution to Radial Problem with Mathematica | |
| Exams | Charge-Dipole Interaction: Derivation of Interaction Energy Expression | NB |
| Exams | How to prepare for the second midterm | |
| Exams | Sample Questions for Midterm 1 (2003) | |
| Exams | Sample Questions for Midterm 2 (2002) | |
| Exams | Midterm 1 Key (Spring 2006) | |
| Poster: | Poster Presentation Guidelines | Link |
Students in the class do not have to purchase the laboratory manual. Each chapter of the lab manual can be downloaded here in the PDF format. Please note that you can follow hyperlinks that are in the PDF files by clicking on the link.
| Experiments | Lab manuals are in the PDF format. Download Adobe Acrobat Here | Acrobat |
| All | Theory Manual for Chem 112L | |
| All | Principles of Biomolecular Recognition | |
| Exp 1: | Conformational analysis of allantoin: Operations Manual | |
| Exp 2 | Enzyme Kinetics | |
| Exp 3 | Protein Unfolding Monitored by Circular Dichroism | |
| Math | Mathematica Tutorial, Part I from Fall Quarter | |
| Math | Mathematica Tutorial, Part II | |
| Math | Mathematica Tutorial, Part III | |
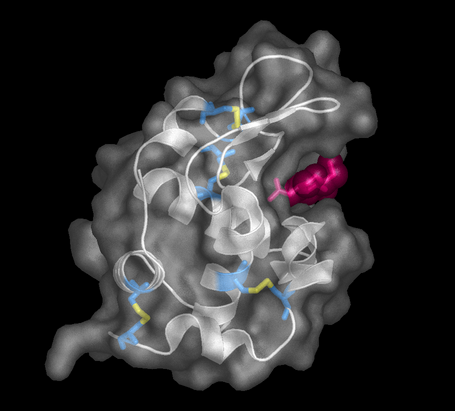
| Exp 4 | Ligand Binding to Lysozyme | |
| Exp 5 | Protein Mass Spectrometry | |
| Exp 6 | Operation Manual for Protein Crystallography |
Data files, instrument manuals, and selected lectures notes to support the course are available below
| Supporting | Support materials PDF format. Download Adobe Acrobat Here | Acrobat |
| Allantoin | Quantum Harmonic Oscillator (using Mathematica) | Link |
| Allantoin | Introduction to Monte Carlo Method (UCSB) | Link |
| Allantoin | Molecular Dynamics (Millisecond Simulations of Proteins, D. E. Shaw et. al.) | |
| Allantoin | Molecular Dynamics (Architecture of Anton, D. E. Shaw et. al.) | |
| Allantoin | Molecular Dynamics (Interview with D. E. Shaw) | |
| Allantoin | Molecular Dynamics (Primer by Furio Ercolessi, University of Udine, Italy) | Link |
| Allantoin | Thorpe-Ingold Acceleration of Oxirane Formation ... (Kostal & Jorgensen) | |
| Allantoin | BOSS User Manual (Jorgensen Research Group at Yale) | Link |
| Allantoin | NMR Lecture Notes | |
| Allantoin | "Structure Determination of Biological Macromolecules in Solution Using NMR Spectroscopy" by Gerhard Wider | |
| Allantoin | "Structure calculation of biological macromolecules from NMR data" by Peter GŁntert | |
| Allantoin | Ab initio calculation of spin-spin coupling constants | Link |
| Allantoin | 1D proton spectrum in Hertz units, sample plot | PNG |
| Allantoin | 1D proton spectrum in ppm units, Full scale, MW2 on 4/09/2012 | |
| Allantoin | 1D proton spectrum in ppm units, Zoom, MW2 on 4/09/2012 | |
| Allantoin | 1D proton spectrum in ppm units, Full scale, TR6 on 4/10/2012 | |
| Allantoin | 1D proton spectrum in ppm units, Zoom, TR6 on 4/10/2012 | |
| Allantoin | 1D proton spectrum in ppm units, Full scale, TR6 on 4/12/2012 | |
| Allantoin | 1D proton spectrum in ppm units, Zoom, TR6 on 4/12/2012 | |
| Allantoin | 1D proton spectrum in Hertz units (MW) | DATA |
| Allantoin | 1D proton spectrum in Hertz units (TR) | DATA |
| Allantoin | Transient NOE spectrum, sample plot | PNG |
| Allantoin | Transient NOE spectrum, irradiate at 5.2 ppm | DATA |
| Allantoin | Transient NOE spectrum, irradiate at 5.8 ppm | DATA |
| Allantoin | Transient NOE spectrum, irradiate at 6.9 ppm | DATA |
| Allantoin | Transient NOE spectrum, irradiate at 8.1 ppm | DATA |
| Allantoin | Transient NOE spectrum, irradiate at 10.5 ppm | DATA |
| Allantoin | COSY (2D) spectrum at 600 MHz | PNG |
| Allantoin | COSY (2D) spectrum zoom in 5-7 ppm | PNG |
| Allantoin | DFQ-COSY (2D) spectrum at 500 MHZ | PNG |
| Allantoin | DFQ-COSY (2D) spectrum at 600 MHz | PNG |
| Allantoin | DFQ-COSY (2D) spectrum at 600 MHz, zoom in 5-7 ppm | PNG |
| Allantoin | NOESY (2D) spectrum at 500 MHz with 0.8 s tMix | PNG |
| Allantoin | NOESY (2D) spectrum at 600 MHz with 0.5 sec tMix (Full) | PNG |
| Allantoin | NOESY (2D) spectrum at 600 MHz with 0.5 sec tMix (Zoom) | PNG |
| Allantoin | NOESY (2D) spectrum at 600 MHz (3D Plot w/ NOE Integrals) | PNG |
| Allantoin | TOCSY (2D) spectrum | PNG |
| Allantoin | Sample NMR Peak Data Files | Singlet Doublet |
| Allantoin | Full 1D spectrum collected in water with WATERGATE | PNG |
| Allantoin | "A correlated ab initio study of Karplus relations ... | |
| Enzymes | "Challenges in Enzyme Mechanism and Energetics" | |
| Enzymes | "Perspective on "Statistical Analysis of Enzyme Kinetics Data"" | |
| Enzymes | "Statistical Analysis of Enzyme Kinetics Data" | |
| Enzymes | "Fitting Enzyme-Kinetic Data to V/K" | |
| Enzymes | "On the Meaning of Km and V/K " | |
| Enzymes | "A Quantitative Approach to Enzyme Inhibition" | |
| Enzymes | "Understanding Enzyme Inhibition" by Raymond Ochs | |
| Enzymes | Optional: "Enzymes and Enzyme Inhibition" by Kalju Kahn | |
| Folding | "Protein Stability Curves" by Becktel and Schellman | |
| Folding | "Heat Capacity in Proteins" by Prabhu and Sharp | |
| Folding | "Synchrotron radiation circular dichroism spectroscopy of proteins ..." | |
| Protein MS | Mass Spectrometry Lecture by Dr. Pavlovich | |
| Protein MS | "Electrospray: Principles and Practice" by Simon Gaskell | |
| Protein MS | "Using Mass Spectrometry for Proteins" by Martha M. Vestling | |
| Protein MS | "Principles of FT-ICR and its Application to Structural Biology" by Barrow et. al. | |
| Protein MS: | "Proteomic Tools For Quantitation By Mass Spectrometry " by Jennie Lill | |
| MS | Mass Spectrometry Data Tables | |
| MS | Mass Spectra Figures 1-3 | |
| MS | Mass Spectra Figures A-F | |
| MS | Mass Spectra For Metabolite Analysis | NB |
| MS | Mass Spectra For Metabolite Analysis | |
| Binding | "Induction of a Remarkable Conformational Change in a Human Telomeric Sequence ... " | |
| Binding | Review of Thermodynamics: Heat Capacity | |
| Binding | Solvatochromism of GFP Chromophore Models | |
| Binding | UV-Vis spectroscopy and Solvatochromism: Comp Chem Notes | Link |
| X-ray | Protein Crystallography: Reading Link | Link |
| X-ray | Crystallography 101 by Bernhard Rupp at LLNL | Link |
| X-ray | "High-resolution neutron protein crystallography ..." | |
| X-ray | Electron Density Map for Crystallography (Coot) | CCP4 |
| X-ray | Electron Density Map for Crystallography (O and Pymol) | OMAP |
| X-ray | Amino Acid Residue Positions for Crystallography | PDB |
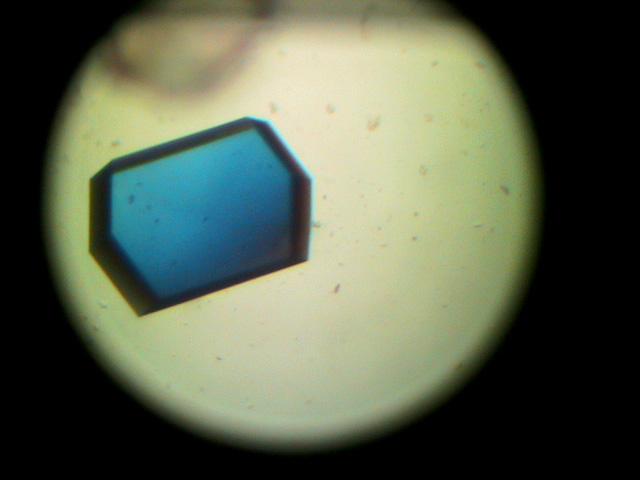
One of the experiments in the Biophysical and Bioanalytical Chemistry Laboratory at UCSB is dedicated to protein crystallography and X-ray diffraction. On the first day, students determine the best conditions for growing lyzozyme crystals via hanging drop crystallization trials. Some of the crystals obtained by students are shown below.


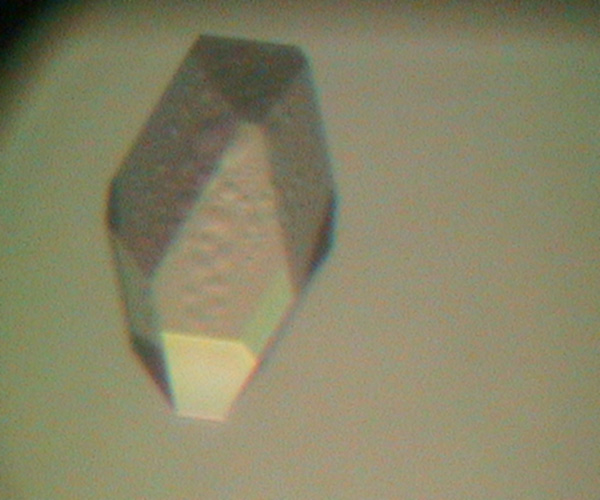
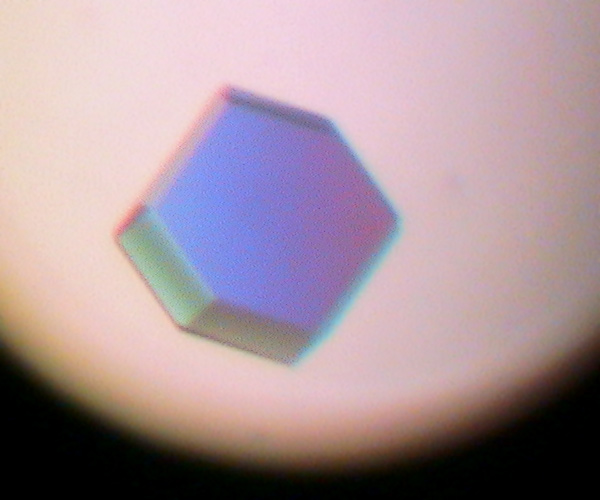
On the following day, crystals were examined and diffraction data was collected on our in-house X-ray diffractiometer. Many students obtained crystals that were large enough for collecting good-quality diffraction data. Some of our best crystals diffracted down to 2.4 Angstroms, which is an acceptable resolution in modern macromolecular crystallography.

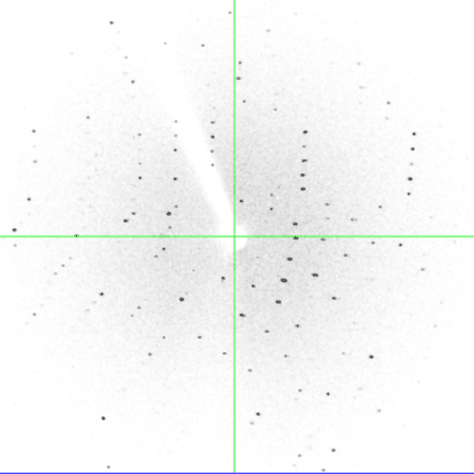
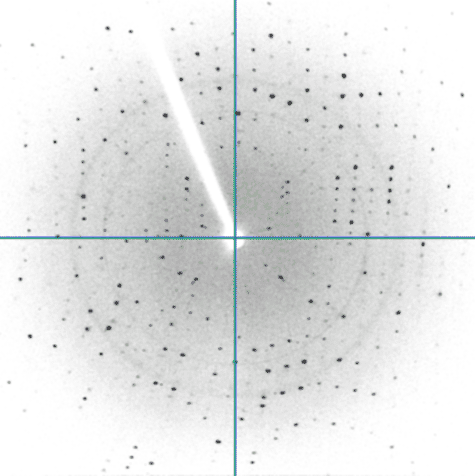
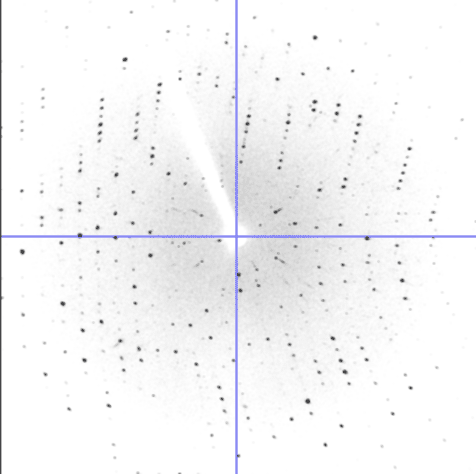
On the last day, students analyzed diffraction data to determine the unit cell characteristics and build a partial protein structure into the electron density map. We thank UCSB's Instructional Development Program for funding the development of this experiment; Dr. Alwyn Jones from Uppsala for providing copies of the program "O", and our gradute student Scott Hauenstein from the Perona group for heping implement this experiment.
Electronic Journals
Protein Data Bank
Wolfram Mathematica Features and Tutorials
PyMOL Molecular Visualization Program
Spectroscopy Online Resources
Protein Prospector: Proteomics Mass Spectrometry Database
UCSB General Catalog
UCSB Gold Login
UCSB Umail Access
UCSB Environmental Health and Safety